Computer scientist Stefano Ceri didn’t know he would end up working on genomics – the study of genes and their functions – until about eight years ago. A professor of database systems at Politecnico di Milano, Ceri was deeply involved in data management research for the first 40 years of his academic career.

An explosion of genomic data, spurred by the advent of next-generation sequencing technologies, led Ceri to become interested in the emerging field of computational genomics. Now, one of his research goals is to use his experience in data management to make the search for genomic information as simple as a Google query. Making this data more accessible could help unlock solutions to illnesses ranging from cancer to COVID-19 by enabling scientists to focus on the important biological questions rather than on the computational steps required to achieve those results.
Ceri’s interest in genomics blossomed when he attended a 2012 scientific meeting hosted at the European Institute of Oncology (EIO), in Milan. “At the time, next-generation sequencing was in its infancy, but it was producing big amounts of data at unprecedented rates. EIO researchers did not know how to manage that data,” he explained.
Following the meeting, Ceri and some of his university colleagues started a collaboration with EIO. They opened PhD opportunities for students with interdisciplinary knowledge to apply data management to genomics.
Genomic information is vast and complex, full of different types of features, or “signals”. These signals include not only mutations, which are changes in the DNA sequence, but also gene expression, a measure of gene activity in specific tissues and conditions (e.g. due to diseases such as cancer), and peaks of expression, which reveal the genomic sites of the DNA where the interaction with a given protein is most significant.
Combining these signals is relevant for answering research questions such as understanding how tumors develop and how they can be cured. (The phrase ‘computational genomics’ emerged in the mid to late 1990s with the availability of complete sequenced genomes.)
When he first became involved with this field, Ceri’s biology background was limited to a high school course. As he was catching up on genomics, he learned that the most recent scientific interest was on understanding signals coming not only from the genes, but also from what is “outside of the genes,” the so-called epigenetics. “It took me several years to scrape the surface of this field,” he said.
Surfing the genome
There are several public genomic data repositories, such as the Encyclopedia of DNA Elements (ENCODE) and The Cancer Genome Atlas (TCGA). Together, these repositories contain an enormous amount of genomic data — but they also presented a challenge. Each of the public data sets is housed separately, use different formats, and use a distinct set of data descriptors.
“My work in the genomic computing area has been focused on building tools to integrate data from various sources, from various formats, into a unique repository where they can be queried [for] a better understanding of global worldwide information,” said Ceri.
Ceri’s first step: integrate and homogenize genomic data from different sources into a single repository hosted at the Polytechnic University of Milan. The second step: make this data easily searchable through user-friendly interfaces that can manipulated by biological science researchers, even if they don’t know how to program.
Thanks to the effort of Ceri’s project – Data-Driven Genomic Computing (GeCo) – researchers from across the globe can now access aggregated genomic data from several sources through a single platform called GenoSurf, which lives on GeCo’s website. The system allows the user to “surf” the genomic data, selecting the properties that are relevant to their research. Then they can visualize and download the results.
Amazon Research Awards issues spring 2021 call for proposals
Proposals related to theory, practice, and novel new techniques are sought in two research areas: Alexa Fairness in AI and AWS Automated Reasoning. The deadline for submissions is April 23.
Along with his colleagues, he also worked to define languages and create tools that can be applied to the repository, making it easier for researchers to identify important regions in the genome sequences, e.g. which genes are mostly expressed in given clinical conditions. This type of complex analysis used to require multiple software tools and data conversions from one software to the next. Ceri’s vision was to give scientists the capability of doing research using a single system that is not only easier to use, but also has more powerful data extraction and analysis capabilities.
“I also developed within my group our own specific data management language for querying those systems, which is called GenoMetric Query Language. It is a new technology and a very powerful and abstract language that can identify genomic regions by combining heterogeneous data – the signals that DNA sends to scientists – thereby making sense of complex phenomena by means of simple computations” Ceri said.
These computations are used to investigate biological questions such as how to assign functions to each portion of the genome, or to understand the genes that could be affected by changes in the genome structure. As computations are heavy, his group became interested in using Amazon Web Services (AWS) as a cloud computing and storage environment. “Our language is built on top of Apache Spark, which is a famous engine for data management computation. And we get the best performance out of Spark by working on the Amazon cloud.”
Ceri decided to apply for the Amazon Machine Learning Research Award (MLRA) when he realized that it was important to have AWS available for his team of PhD students and other associated researchers. His 2019 MLRA award allowed the group to use AWS in different ways. That included demonstrating the scalability of systems developed by the group, which require speed-up and scale-up experiments that involve progressively more nodes in the AWS cloud.
Investigating viral sequences
GenoSurf was already being used by researchers in Italy and other countries, mainly in oncology studies, when the COVID-19 pandemic started. Since many of the collaborators were hospitals, the projects were temporarily suspended, as they concentrated on handling the health crisis.
The GeCo project also redirected its efforts to the study of viral genome sequences. Ceri’s team used their expertise from GenoSurf to develop ViruSurf. This search engine aggregates data from viral genome sequences stored on different databases. Any researcher can access the system and perform queries such as when a given mutation appeared for the first time and how it is spreading.
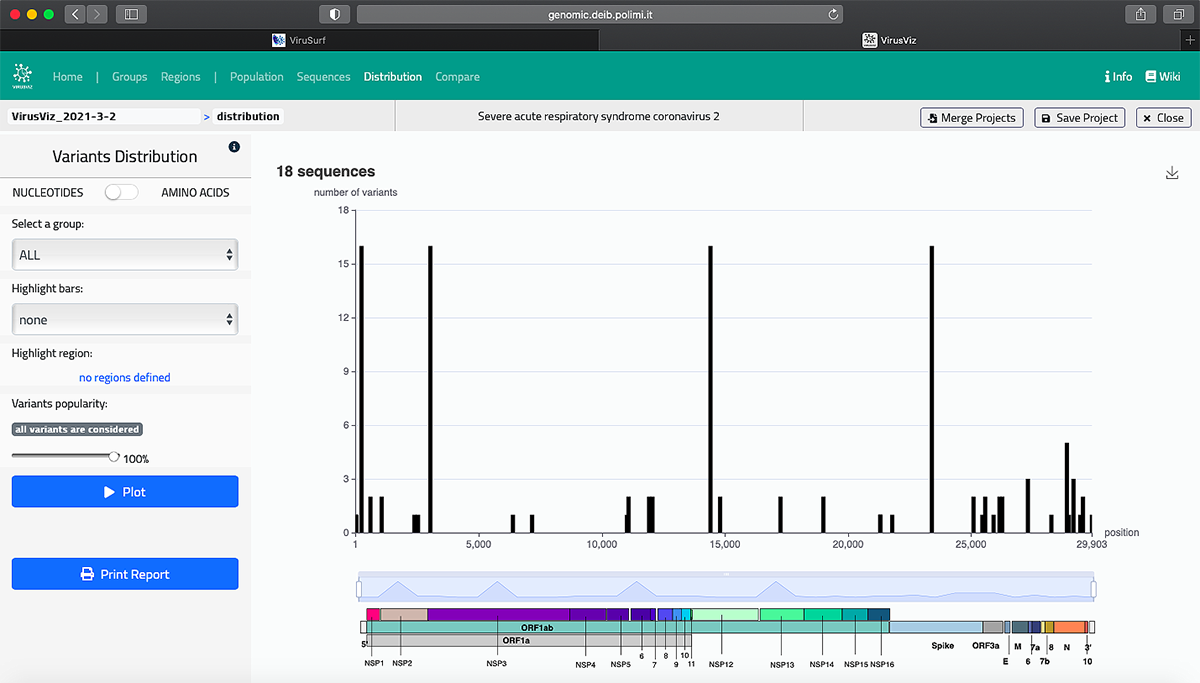
The system is constantly updated to include all the sequences that have been produced of SARS-CoV-2 around the world. At the moment, there are about 650,000 of them. “For data import and curation, including variant search, we use algorithms and tools that are heavy in terms of computation. That's where AWS comes in again and helps us do effective and fast computation,” Ceri said.
When the pandemic recedes, Ceri hopes to complete a few projects that have been on pause as hospitals cope with COVID-19 patients. These initial collaborations are on prostate cancer prevention and on precision medicine for ovarian cancer and Hodgkin lymphoma. For someone who, as early as eight years ago, thought of the DNA simply as a “four-letter encoding,” Stefano Ceri is creating a marker for himself in genetics research.